We provide genomic insights for your patients.
What is the intended use of Summit 2.0 Comprehensive Genomic Profile?
Summit 2.0 interrogates tumor-derived nucleic acid in CSF to help inform the diagnosis and management of primary and secondary CNS malignancies.
What does the Summit 2.0 test evaluate?
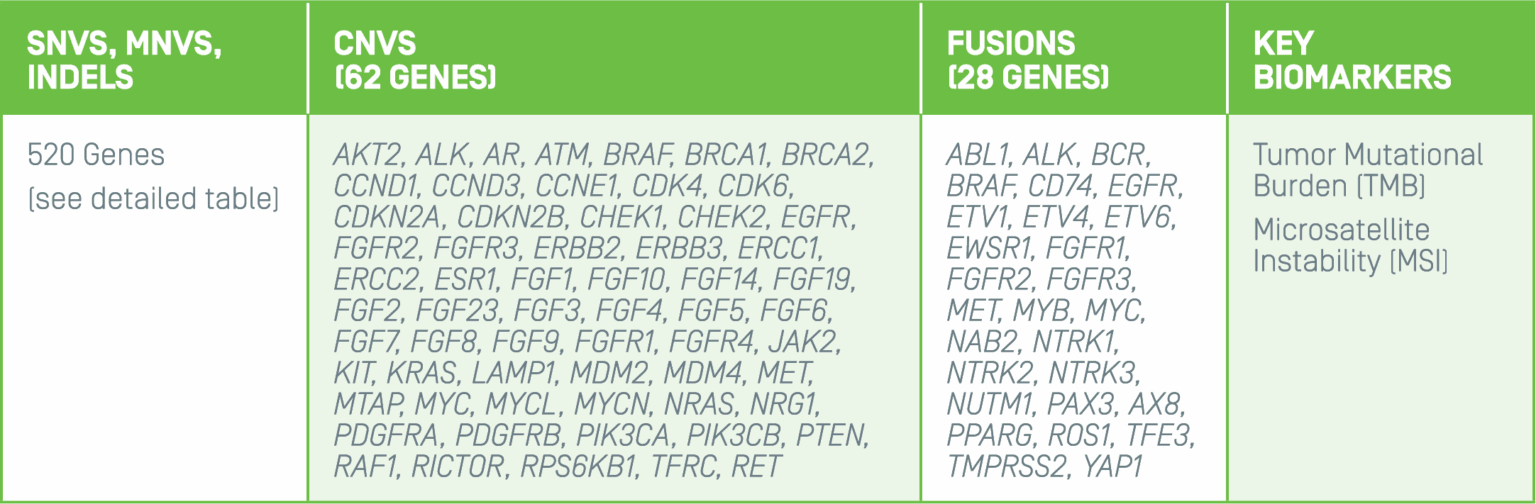
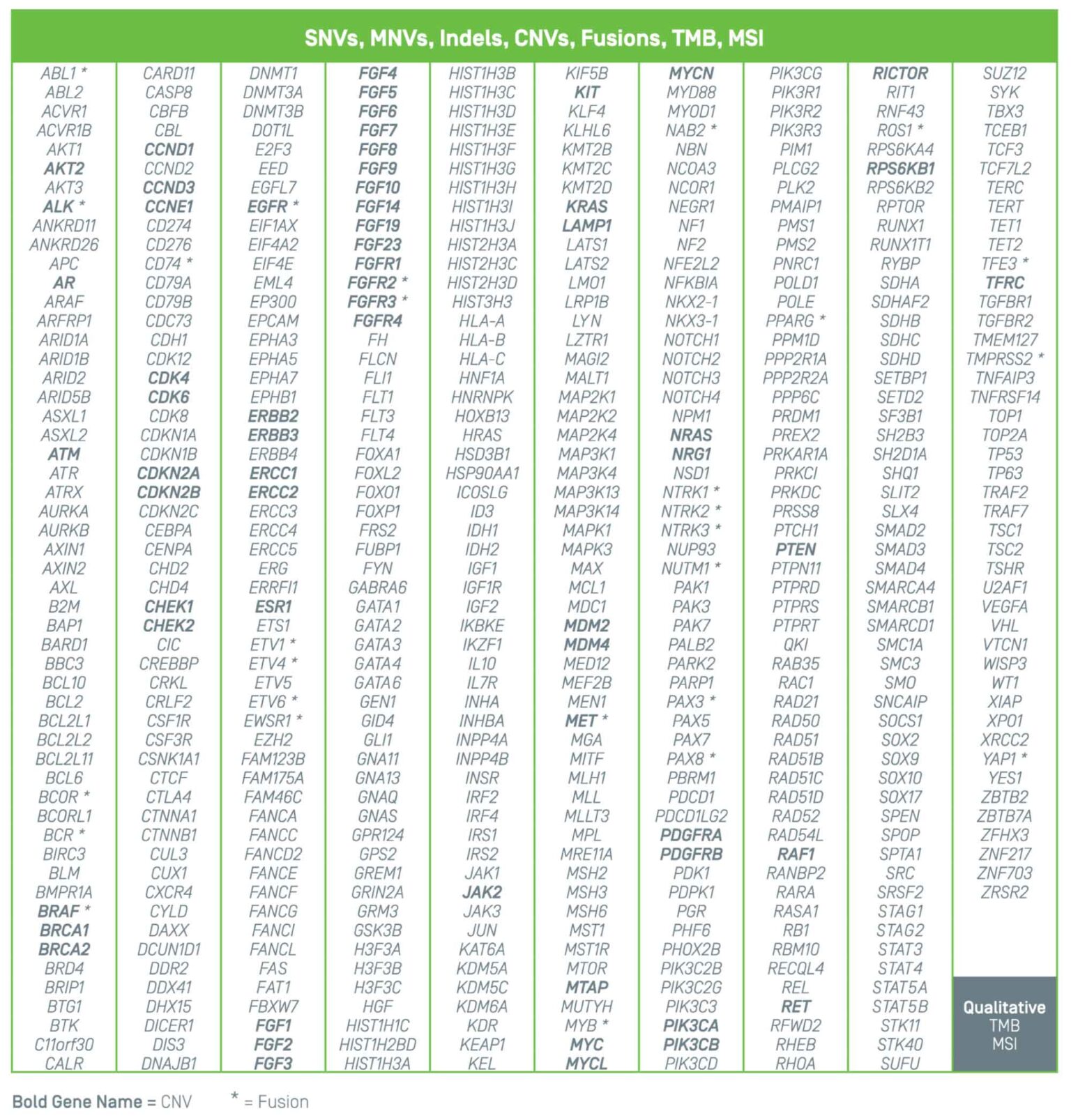
Summit 2.0 investigates cerebrospinal fluid (CSF) for clinically relevant variants known to be associated with brain and spinal cord cancers. The 500+ gene panel includes single nucleotide variants, multi-nucleotide variants, insertions, deletions, copy number variants, chromosomal arm level loss/gain, fusions, and the key biomarkers, TMB and MSI.
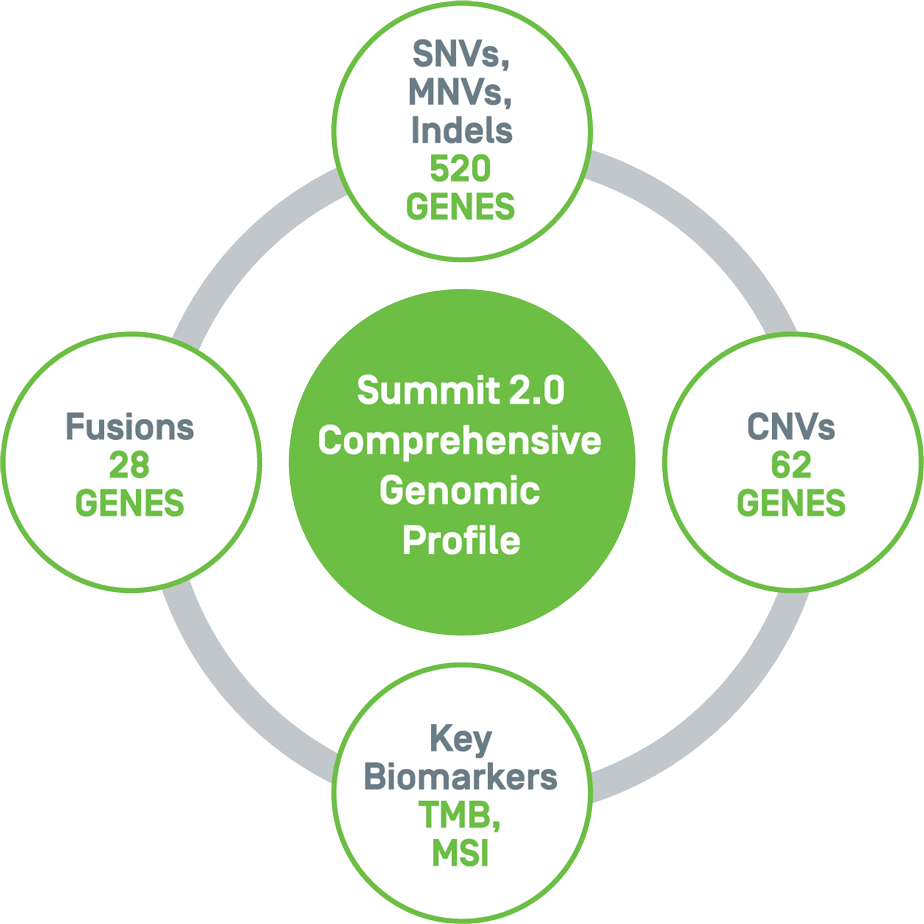
Large-scale genomic profile addresses clinical needs
To learn more about tumor derived DNA in CNS tumors, click or tap on this link to see the lecture, “Applications of CSF Tumor DNA as a Biomarker for CNS Tumors” as presented at SNO/ASCO 2025.
What technology is used for performing Summit 2.0?
The methodology involves next generation sequencing, proprietary duplex sequencing, and low pass whole genome sequencing. Post target enrichment, libraries are sequenced on the Illumina® NovaSeq XPlus. Sequencing data is processed through custom bioinformatics pipelines. Variants are called against the human genome build reference hg19.

What is the intended use of Vantage?
Vantage evaluates MGMT promoter methylation status in cerebrospinal fluid (CSF). This test provides prognosis in high grade glioma patients. With positive methylation of MGMT, this corresponds to better therapeutic response to alkylating agents (e.g. temozolomide). MGMT promoter methylation testing is recommended in all high-grade gliomas. Since MGMT status can change, this testing is a significant advancement in helping treat these patients. Vantage can be ordered concurrently with Summit 2.0 or independently.
What does the Vantage test evaluate?
Vantage test uses quantitative polymerase chain reaction methodology followed by high-resolution melt analysis to evaluate MGMT (O-6-methylguanin-DNA methyltransferase) promoter methylation status in tumor derived DNA extracted from cerebrospinal fluid (CSF) of known or suspected central nervous system tumors. The Vantage test reports a qualitative result and is interpreted as “Positive – Methylated,” “Negative – Unmethylated” or “Indeterminate – Results Equivocal.”
What methodology is used for performing Vantage?
The methodology optimizes performance on enzymatically converted tumor-derived nucleic acid in CSF which minimizes DNA damage.1 It targets 12 cytosine-phosphate-guanine (CpG) island sites (#72-83) within exon 1 of the MGMT gene. The assay uses qPCR (quantitative polymerase chain reaction) followed by high-resolution melt analysis using the EpiMelt MGMT assay (MethylDetect®). Methylated and unmethylated melting temperature peaks are evaluated. Specimens with results above the validated 25% methylated control are interpreted as “Positive – Methylated.” When a peak detected aligns with the control that is unmethylated, results are reported as “Negative – No MGMT methylation detected.” Specimens with results between unmethylated and methylated control are interpreted as “Indeterminate – Results Equivocal.” Vantage testing can be performed on the same specimen as Summit 2.0.
How can we learn more about the Summit 2.0 and Vantage tests?
Belay Diagnostics technical bulletins are available for download:
Additionally, here below are links to recent webinars on the subject:
Can we obtain data files?
Belay Diagnostics does not provide raw data files.
Reference
1. New England Biolabs, Inc. NEBNext® Enzymatic Methyl-seq (EM-seq™). Updated April 2019. Accessed May 20, 2024.


